microRNAs – Web-based tools
You need more information about “your” microRNAs? View our collection of over 20 resources and find your favorite one.
microRNA target prediction tools
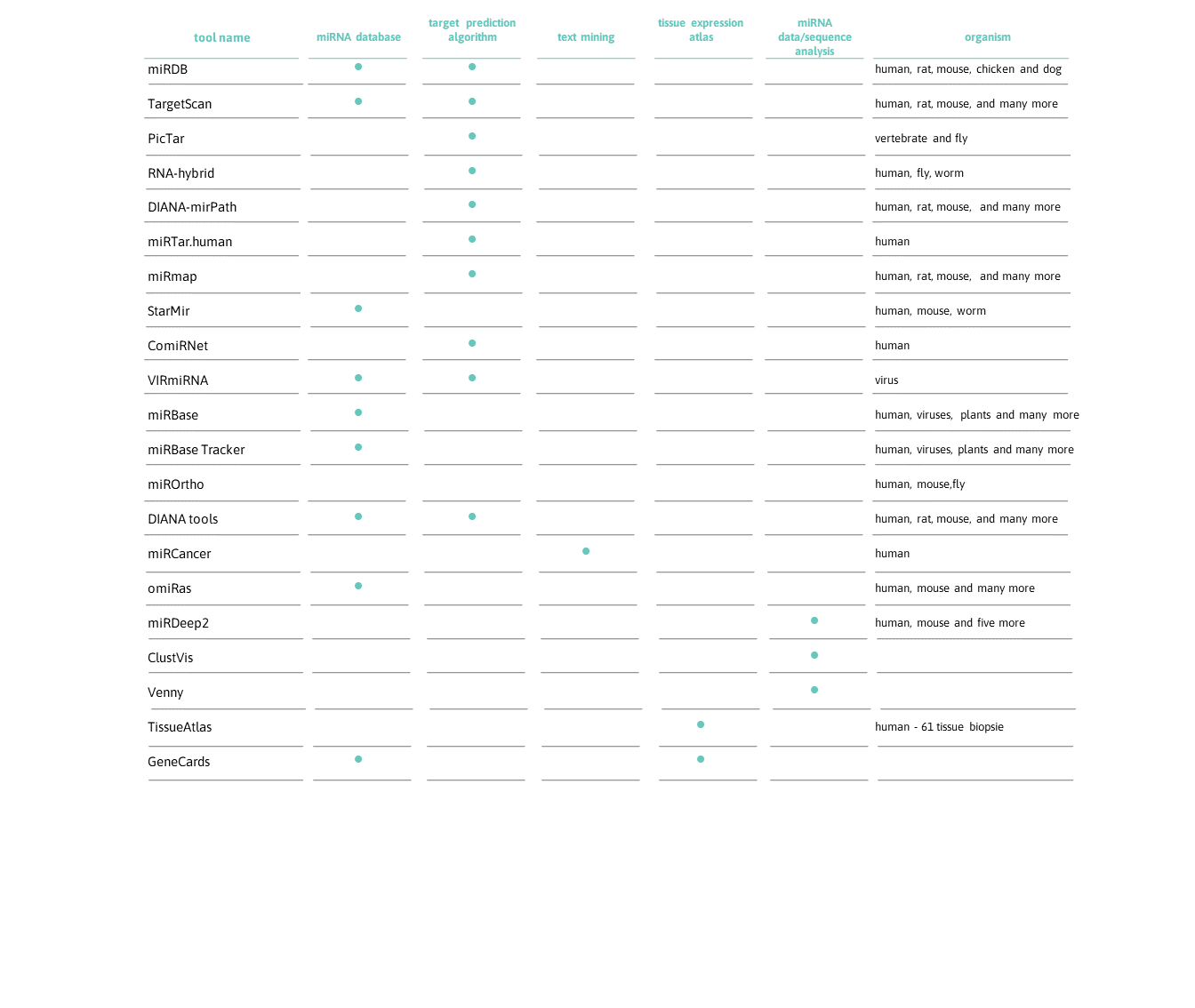
miRDB is an online database for miRNA target prediction and functional annotations. All the targets in miRDB were predicted by a bioinformatics tool, MirTarget, which was developed by analyzing thousands of miRNA-target interactions from high-throughput sequencing experiments. Common features associated with miRNA target binding have been identified and used to predict miRNA targets with machine learning methods. miRDB hosts predicted miRNA targets in five species: human, mouse, rat, dog and chicken.
TargetScan predict regulatory targets of vertebrate microRNAs (miRNAs) by identifying mRNAs with conserved complementarity to the seed (nucleotides 2-7) of the miRNA. An over-representation of conserved adenosines flanking the seed complementary sites in mRNAs indicates that primary sequence determinants can supplement base pairing to specify miRNA target recognition.
PicTar is a computational method for identifying common targets of microRNAs. Statistical tests using genome-wide alignments of eight vertebrate genomes, PicTar’s ability to specifically recover published microRNA targets, and experimental validation of seven predicted targets suggest that PicTar has an excellent success rate in predicting targets for single microRNAs and for combinations of microRNAs.
RNA-hybrid predicts multiple potential binding sites of miRNAs in large target RNAs. In general, the program finds the energetically most favorable hybridization sites of a small RNA in a large RNA.
DIANA-mirPath can utilize predicted miRNA targets (in CDS or 3’-UTR regions) provided by the DIANA-microT-CDS algorithm or even experimentally validated miRNA interactions derived from DIANA-TarBase. These interactions (predicted and/or validated) can be subsequently combined with sophisticated merging and meta-analysis algorithms.
miRTar is a tool that enables easily to identify the biological functions and regulatory relationships between a group of known/putative miRNAs and protein coding genes. It also provides perspective of information on the miRNA targets on alternatively spliced transcripts.
Includes experimentally verified miRNAs and their targets, also contains RNAhybrid and TargetScan target prediction module.
Software for statistical folding of nucleic acids and studies of regulatory RNAs based on CLIP data (sequence, thermodynamic and target structure features).
ComiRNet uses 10 miRNA target prediction databases, stores approximately 5 million predicted interactions between 934 human miRNAs and 30,875 gene transcripts (mRNAs) which are exploited in the construction of the hierarchies of overlapping biclusters representing potential miRNA regulatory networks.
VIRmiRNA is the first dedicated resource on experimental viral miRNA and their targets. VIRmiRNA is a resource for experimentally validated viral miRNA and its target reported in literature. It comprises of three sub-databases-VIRmiRNA, VIRmiRtar and AVIRmir. VIRmiRNA contains 1308 miRNA entries encoded by 44 viruses.
microRNA databases
The miRBase database is a searchable database of published miRNA sequences and annotation. Each entry in the miRBase Sequence database represents a predicted hairpin portion of a miRNA transcript (termed mir in the database), with information on the location and sequence of the mature miRNA sequence (termed miR). Both hairpin and mature sequences are available for searching and browsing, and entries can also be retrieved by name, keyword, references and annotation. All sequence and annotation data are also available for download.
An easy-to-use online database that keeps track of all historical and current miRNA annotation present in the miRBase database.
miROrtho contains predictions of precursor miRNA genes covering several animal genomes combining orthology and a Support Vector Machine. It provides homology extended alignments of already known miRBase families and putative miRNA families.
DIANA-TarBase v8.0 is a reference database devoted to the indexing of experimentally supported microRNA (miRNA) targets. Its eighth version is the first database indexing >1 million entries, corresponding to ∼670 000 unique miRNA-target pairs. The interactions are supported by >33 experimental methodologies, applied to ∼600 cell types/tissues under ∼451 experimental conditions. It integrates information on cell-type specific miRNA-gene regulation, while hundreds of thousands of miRNA-binding locations are reported.
miRCancer : microRNA Cancer Association Database
miRCancer provides comprehensive collection of microRNA (miRNA) expression profiles in various human cancers which are automatically extracted from published literatures in PubMed. Search by cancer names or the on-site sequence analysis tools is possible.
The new version of miRWalk stores predicted data obtained with a maschine learning algorithm including experimentally verified miRNA-target interactions. The focus lies on accuracy, simplicity, user-friendly design and mostly up to date informations.
omiRas provides an interface to generate a miRNA-mRNA interaction network for user selected microRNAs. Interactions between genes and miRNAs are determined by the interesection of 7 databases (TargetScan, miRanda, PICTAR, PITA, miRDB, TarBase, miRConnect).
microRNA deep sequencing tools
miRDeep2 is a completely overhauled tool which discovers microRNA genes by analyzing sequenced RNAs.
microRNA data analyses tools
This web tool allows users to upload their own data and easily create Principal Component Analysis (PCA) plots and heatmaps.
An interactive tool for comparing lists using Venn Diagrams with support for up to 4 sets.
miRNA tissue expression databases
TissueAtlas allows identifying the tissues of origin of miRNAs and provides insights into specificity and heterogeneity of miRNAs with respect to tissues.
RATEmiRs is a database of rat microRNA-Seq data in several tissues. “Data Driven” pipelines are available to detect tissue-specific and tissue-enriched microRNAs. “User Driven” entrez is available to enter a list of IDs of mature microRNAs. Organ-specific pipelines detects microRNAs that are specific for a particular set of tissues that comprise of an organ.
GeneCards is a searchable, integrative database that provides comprehensive, user-friendly information on all annotated and predicted human genes. It automatically integrates gene-centric data from ~125 web sources, including genomic, transcriptomic, proteomic, genetic, clinical and functional information.
The GeneCards Suite of Databases
- MalaCards: A database of human maladies and their annotations
- PathCards: The integrated database of human pathways and their annotations
- GeneALaCart: Generates a file of GeneCards annotations for your list of genes
- GeneLoc: Presents an integrated map for each human chromosome, based on data integrated by the GeneLoc algorithm